MGTdb website features
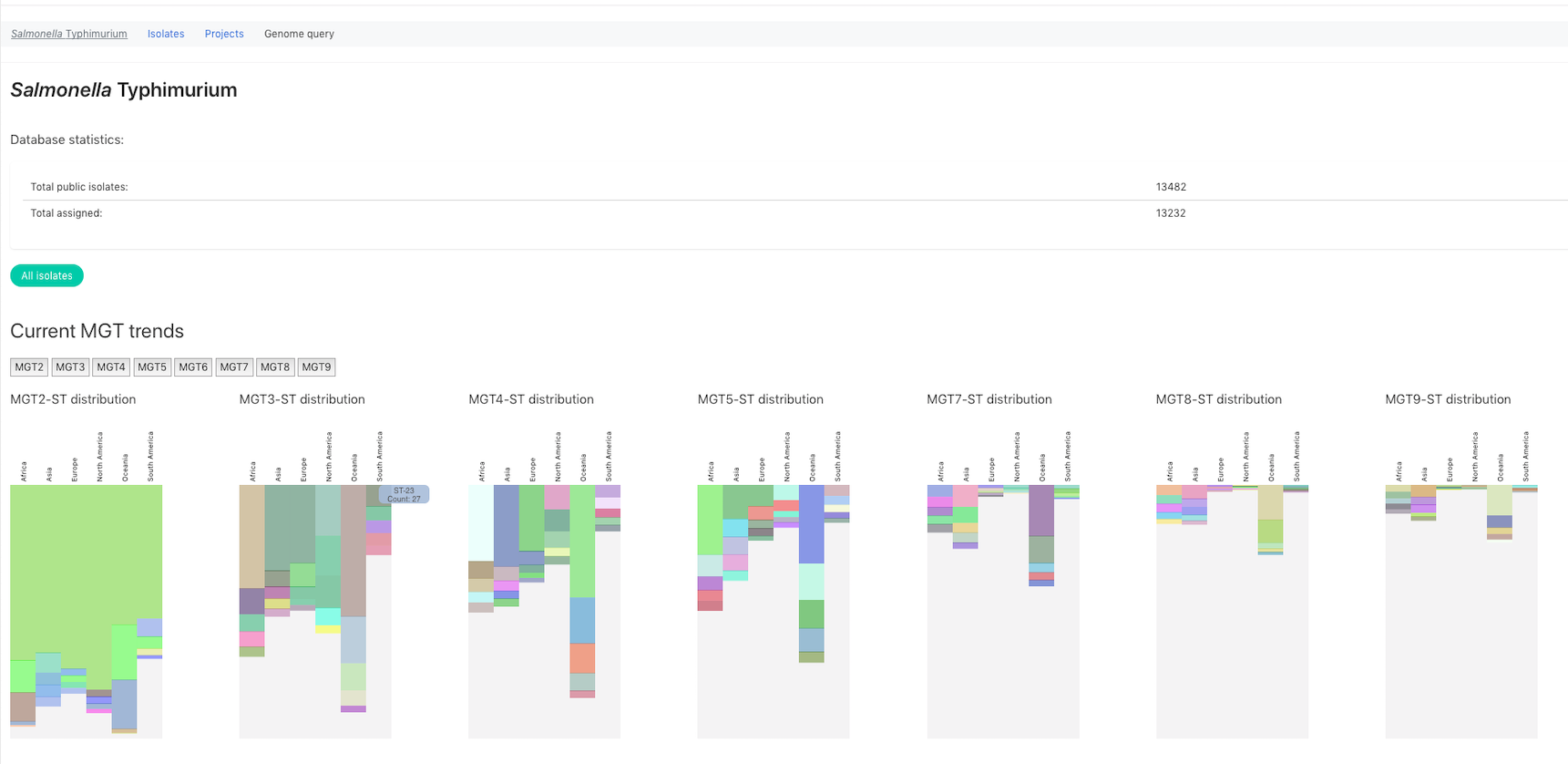
MGTdb is a webservice and database system which enables a user to upload isolates, obtain MGT assignments and explore these in the context of the local and global database of isolates. MGTdb is currently available for three organisms namely, Salmonella Typhimurium (Payne et al. 2020) at https://mgtdb.unsw.edu.au/salmonella, Salmonella Enteritidis (Luo et al. 2021) at https://mgtdb.unsw.edu.au/enteritidis and Vibrio cholerae (Cheney et al. 2021) at https://mgtdb.unsw.edu.au/vibrio.
The MGT website is available at: https://mgtdb.unsw.edu.au
Account features
You can create an account with MGT which enables you to submit isolates, and get an MGT assignment for them.
Creating an account
You can set up an account at the website. Simply click on Register at the top navigation bar, and enter in your details.
Doing so will send you an email with a link (check your junk email if you don’t receive an email within a few minutes). Click or copy and paste the link, and your account will be active, and you can log in with your supplied details.
Note: Certificates will be added soon to make your communication with the website httpsecure.
Uploading isolates
Once you log in, you can add projects, and isolates to a project.
Creating a project is straightforward. Simply click on ‘Projects’ in the top navigation bar. Then select the organism you want to create a project for.
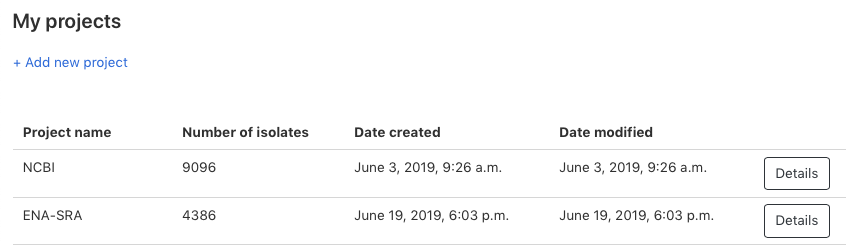
Clicking on ‘+ Add new project’ to create a new project. Only a project name is required to create a project.
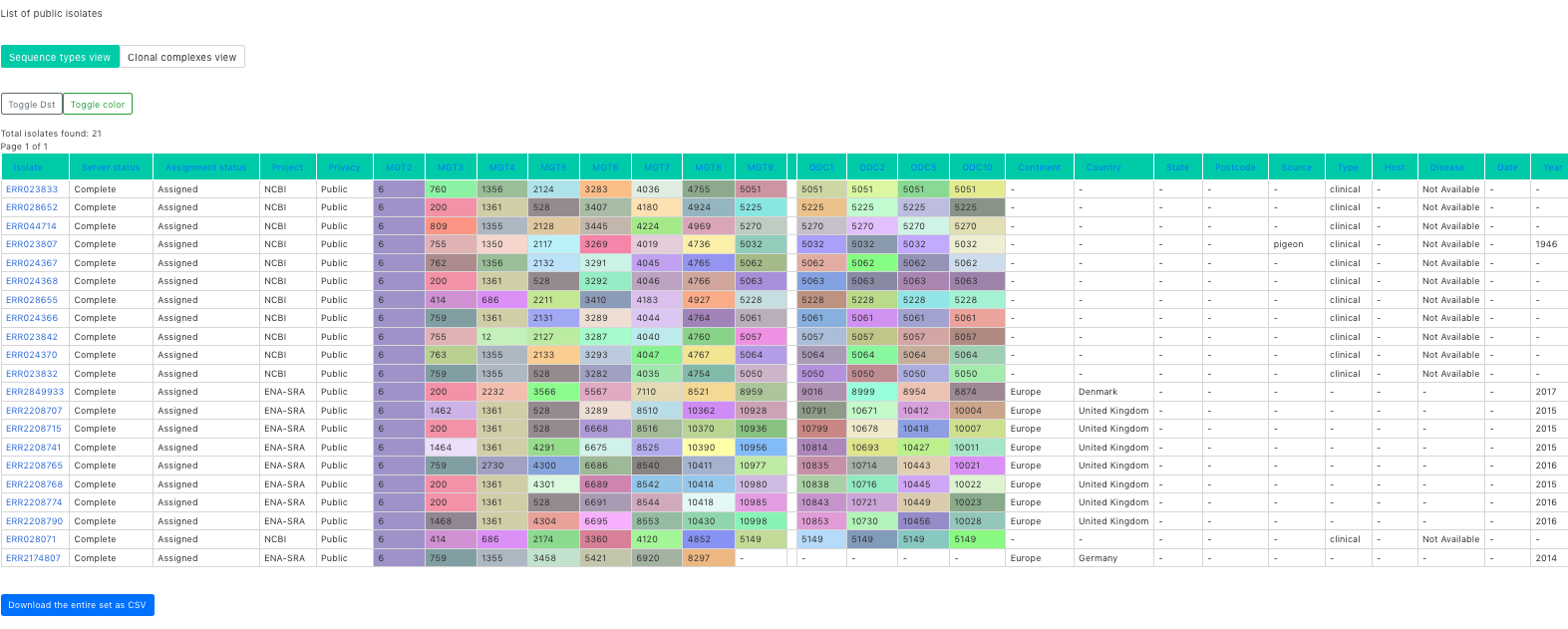
Once a project is created, navigate to the project detail. Then clicking on ‘+ Upload new isolate’ enables you to add isolates.
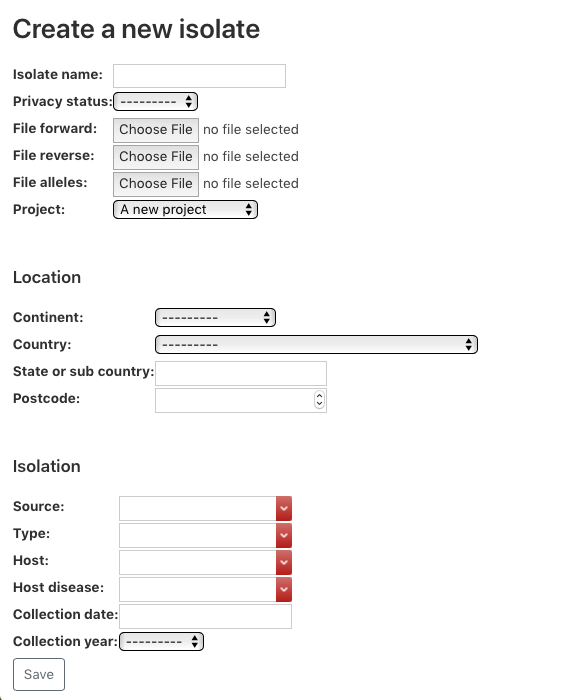
Webpage to enable adding isolate to the MGT database. The relevant information can be supplied here.
Note, that for the files to be uploaded, either Illumina sequenced files forward and reverse should be supplied, or alleles files. The advance of providing alleles file is that the uploaded file is a lot smaller (if internet speeds are an issue).
To generate the allele files locally see section Generating alleles file locally. Apart from the uploaded files, Collection year, Country, Countinent, Privacy status and Isolate name are compulsory fields.
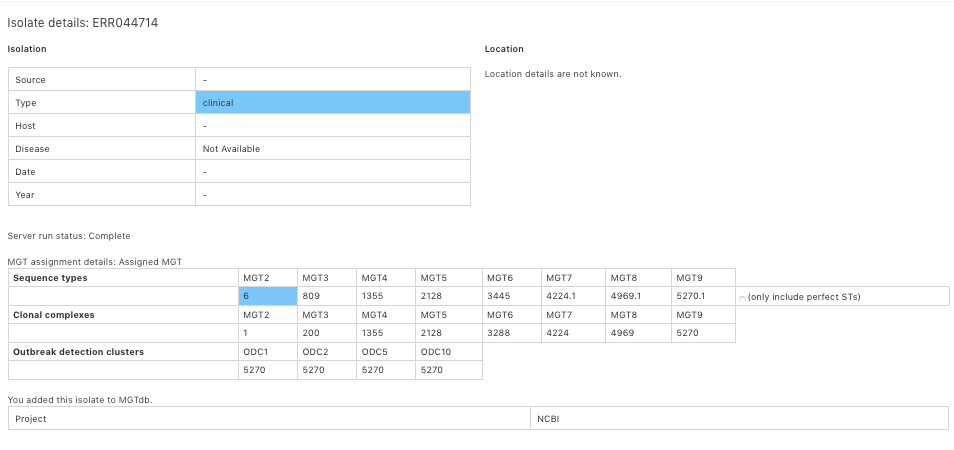
Once information is received on the server, it is submitted as a job, where the alleles are first extracted (if reads are provided), following which an MGT assignment is made and added to the database. Details about this process can be found on Analysis pipeline.
Once an MGT assignment is made, an email notification is sent.
Privacy
We take your uploaded isolates privacy very seriously. Your isolates are made public only if you specify. Furthermore, if you delete your isolates, then all associated isolate meta-data and the uploaded files are deleted.
Note that when a project is deleted, all the isolates in that project and their associated data are deleted.
References
Cheney L, Payne M, Kaur S, Lan R. Multilevel Genome Typing Describes Short-and Long-Term Vibrio cholerae Molecular Epidemiology. Msystems. 2021 Aug 24;6(4):e00134-21.
Luo L, Payne M, Kaur S, Hu D, Cheney L, Octavia S, Wang Q, Tanaka MM, Sintchenko V, Lan R. Elucidation of global and national genomic epidemiology of Salmonella enterica serovar Enteritidis through multilevel genome typing. Microbial genomics. 2021 Jul 22;7(7):000605.
Payne M, Kaur S, Wang Q, Hennessy D, Luo L, Octavia S, Tanaka MM, Sintchenko V, Lan R. Multilevel genome typing: genomics-guided scalable resolution typing of microbial pathogens. Eurosurveillance. 2020 May 21;25(20):1900519.